Every day, clinicians must make life-or-death decisions based on incomplete or scattered information. Imagine an emergency room doctor trying to decide if a patient with chest pain should be admitted for observation or sent home. Or a care team trying to predict whether a recently discharged patient might be readmitted within 30 days.
These decisions carry enormous risks, and while artificial intelligence (AI) models are beginning to assist, they may often fall short. They can sound persuasive, but there are risks for models to make mistakes, invent facts, miss critical medical details, or fail to focus on the information that truly matters for a patient’s outcome. KARE1, the Knowledge-Augmented Retrieval Engine, is developed to help solve this problem—by helping AI reason more like experienced clinicians and retrieve the precise knowledge needed for each case. KARE introduces innovations such as multi-source knowledge structuring, dynamic knowledge retrieval, and a reasoning-enhanced prediction framework. These enable KARE to provide more accurate predictions and explanatory chains that justify each decision, aligning closely with clinical reasoning.
The challenge
Large language models (LLMs) have shown impressive abilities in understanding and generating human-like text. However, in healthcare settings, where accurate predictions can guide interventions and save lives, LLMs need to achieve high level of accuracy and reliability. In the medical context it is important that models don’t fabricate information, a phenomenon known as hallucination, or fail to grasp subtle, domain-specific cues that are essential in medicine.
To improve reliability, researchers have turned to Retrieval-Augmented Generation (RAG), which supplements LLMs with external knowledge. But not all retrieval is helpful2. If the documents retrieved are too broad, semantically close but clinically irrelevant, or not aligned with the prediction task, the model’s performance can actually degrade.
Take the case of heart failure prediction. A generic retrieval engine might surface papers on “ischemic heart disease” because the text is similar. But unless it specifically includes markers like left ventricular ejection fraction (LVEF) or NT-proBNP levels, two key indicators used in diagnosing and managing heart failure, is likely not to be clinically actionable.
Enter KARE
To bridge this gap, researchers from GE HealthCare and the University of Illinois Urbana-Champaign (UIC) developed KARE, a knowledge graph–based framework that strengthens language model reasoning with structured, clinically relevant data.
KARE does not simply pull up more documents. It builds a detailed, multi-source medical knowledge graph by integrating structured biomedical databases, such as the Unified Medical Language System (UMLS), peer-reviewed clinical literature, such as PubMed, and insights generated by LLMs. It then applies a technique called hierarchical community detection, a method for organizing large graphs into meaningful clusters, to uncover related groups of medical concepts. These clusters, or “communities,” are enriched with language model–generated summaries, helping surface high-signal, focused knowledge.
The result is a system where, when a clinician or model needs to predict something like hospital readmission, KARE brings forward the most relevant insights grounded in real-world patient care, not just textually similar content.
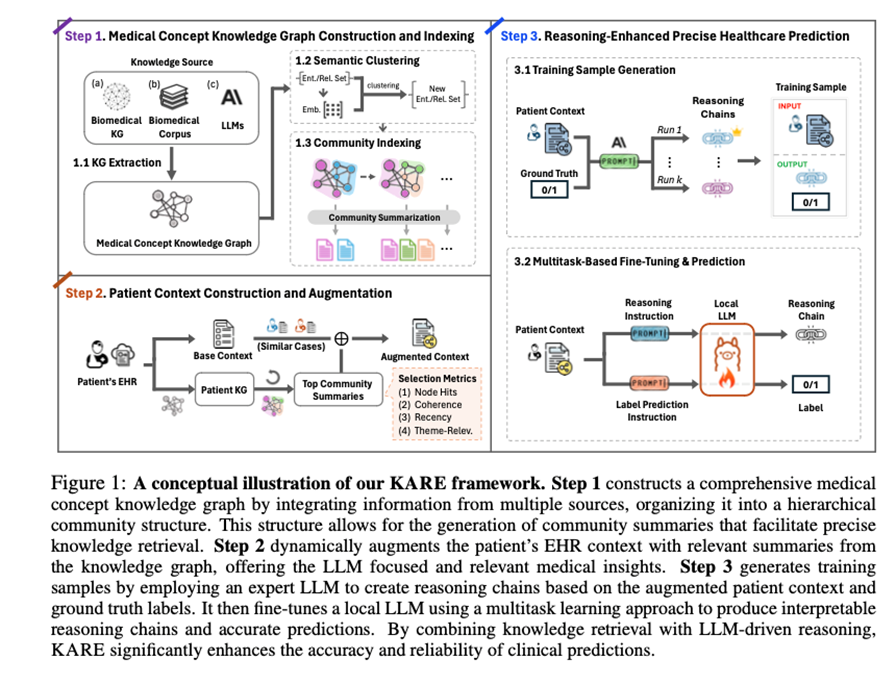
Three new innovations
KARE introduces three innovations that move beyond traditional search-and-retrieve methods:
- Multi-Source Medical Knowledge Structuring
KARE combines multiple knowledge streams into a unified foundation. These include:- Biomedical ontologies like UMLS, which organize medical terms and their relationships,
- Peer-reviewed publications from databases like PubMed, which provide evidence-based findings,
- LLM-generated associations that extend the graph with inferred insights.
For example, for suspected pulmonary embolism, KARE integrates data on D-dimer levels from literature, thrombotic risk pathways from UMLS, and potential links between recent surgery and clot formation, resulting in a richer, clinically precise picture.
- Dynamic Knowledge Retrieval
Unlike static databases, KARE tailors its retrieval to the patient at hand. Its algorithm, called Dynamic Graph Retrieval and Augmentation (DGRA), looks at a patient’s clinical data and selects the most applicable subset of the graph. It uses measures such as concept overlap, relevance, recency, and coherence to guide retrieval.
In congestive heart failure, for instance, KARE retrieves information focused on ejection fraction, renal function, and diuretic response, bypassing irrelevant cardiovascular material.
- Reasoning-Enhanced Prediction Framework
KARE enables step-by-step reasoning in its predictions. It uses powerful LLMs, such as Claude 3.5 Sonnet, to generate explanatory chains that justify each decision. These chains are used to train a more efficient model, such as Mistral-7B-Instruct-v0.3, which can then produce either a full reasoning trace or just a final prediction.
Imagine a readmission risk prediction that not only tells you the risk is high, but walks you through how comorbid diabetes, a recent discharge, missed follow-ups, and abnormal lab results contributed to that assessment. That is the kind of transparency clinicians need.
Implications and real-world impact
KARE was evaluated using two large, anonymized datasets of ICU patients3, MIMIC-III and MIMIC-IV. Across both, it significantly outperformed standard models:
- Mortality Prediction: KARE achieved up to a 15.0% improvement in predictive accuracy.
- On MIMIC-III: 24.7% sensitivity,
- On MIMIC-IV: 73.2% sensitivity.
- Readmission Prediction: KARE achieved up to a 12.7% increase in accuracy.
- Macro F1 score on MIMIC-III: 73.7%,
- Macro F1 score on MIMIC-IV: 73.8%.
These improvements translate into real clinical value. Greater sensitivity means fewer high-risk patients slip through unnoticed. Better F1 scores could help alert doctors to true positives while reducing overload with false alarms.
Beyond the numbers, KARE’s transparency could give clinicians something far more valuable: trust. A prediction system that explains itself, shows its sources, and aligns with a doctor’s clinical reasoning is far more likely to be adopted and acted upon.
Conclusion
Healthcare needs AI systems that do not just predict but explain. Systems that understand the difference between sounding smart and being clinically useful. KARE takes a significant step in that direction by aligning LLM capabilities with structured medical knowledge and real-world clinical logic.
Imagine a future where discharge planning is proactive, more tailored to each patient’s unique risks. Imagine a future where AI flags possible complications before symptoms arise, where rare diseases do not go undiagnosed for years. That is the kind of future KARE helps make possible.
This is just the beginning. Future research will explore how KARE can adapt to more complex cases, such as integrating medical imaging, genomics, or wearable data, and how it might support diagnosis in rare or poorly understood conditions.
To dive deeper into KARE’s design and evaluation, explore the full paper: https://arxiv.org/abs/2410.04585.
By uniting biomedical knowledge graphs with LLM reasoning, KARE is building a bridge between machine intelligence and medical intuition, one interpretable prediction at a time.
- Concept only. This work is in concept phase and may never become a product. Not for Sale. Any reported results are preliminary and subject to change. Not cleared or approved by the U.S. FDA or any other global regulator for commercial availability. ↩︎
- https://arxiv.org/abs/2505.08445 ↩︎
- https://arxiv.org/abs/2410.04585 ↩︎